Features
- Searching for monophyletic relationship among groups of taxa.
- Filtering by bootstrap support values associated with the monophyletic clades.
- Filtering by tree complexity (number of taxa in a tree).
- Filtering by family complexity (number of genes per taxon in a tree).
- Clustering trees (genes) into tree clusters (gene families).
- Grouping OTUs using a taxonomy reference tree.
- Clean and reusable implementation of common procedures to manipulate tree data structure such as Newick parsing, tree traversal and rerooting.
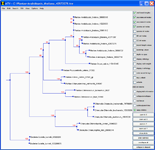
- Visualizing tree topology using A Tree Viewer (ATV).
Usage
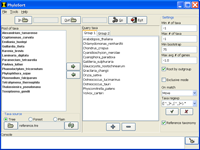
Input
- Select an input (readable) folder for the input trees. Once the input folder is selected, all trees will be scanned to retrieve the set of all taxa in all trees. These taxa will show in the list of available taxa on the left hand side of the main window.
- Select an output (writeable) folder for the matching trees.
- Define query taxa group(s) by moving taxa from the list of available taxa to the query taxa on the right hand side of the main window. Groups can be added and removed using the plus (+) and minus (−) buttons, respectively.
Settings
Mode
PhyloSort offers two search modes, inclusive (default) and exclusive:
- In the exclusive mode, any taxa from the query group(s) exist in the tree, will have to be located within the monophyletic clade in order to qualify that clade and consider the tree as a match tree.
- In the inclusive mode, the restriction of the exclusive mode is relaxed such that a "qualifying" monophyletic clade is accepted regardless whether there are additional taxa from the query taxa located somewhere else in the tree outside that clade.
Root
PhyloSort searches for an outgroup and reroots the tree on the identified outgroup. If an outgroup could not be found (i.e. all taxa in the tree are subset of the query taxa), the tree is searched without rerooting.
Filtering
The following is a list of filtering parameters that could be used to adjust the matching output trees (any of these filters can be turned off by setting its value to a negative number).
- Minimum number of taxa: the least number of taxa must be contained in a tree. (integer number)
- Maximum number of taxa: the largest number of taxa a tree can contain. (integer number)
- Minimum bootstrap support: the minimum bootstrap support value on the monophyletic clade. (decimal number)
- Maximum average number of genes (or copies) per taxon. (decimal number)
All the settings and filters can be loaded from a configuration file, which may include an item or more to configure. The configuration file is a key-value-pair-based text file. In the GUI interface, a configuration file can be loaded via the menu "File" → "Load configuration" (or shortcut Ctrl+O). In the command line interface, it can be loaded through the system environment phylosort.config (e.g. -Dphylosort.config=filename). The following list shows all configurable properties and their possible values.
phylosort.root.outgroup:yes(default) ornophylosort.mode:inclusive(default) orexclusivephylosort.minimum.number.taxa: integer numberphylosort.maximum.number.taxa: integer numberphylosort.minimum.bootstrap.support: decimal numberphylosort.maximum.average.number.copies: decimal numberphylosort.on.match.action:copy,moveorcount(default)phylosort.pattern: regular expression
Output
Based on the selection criteria and the phylosort.on.match.action, trees with matching monophyletic clades are counted, copied or moved from the input folder to the output folder.
Command line
Command line arguments
Graphical user interface
Prerequisites
PhyloSort requires that Java Runtime Environment (JRE) version 1.5 or later is installed. JRE can be downloaded at java.com.
Licenses
The source code is licensed under The GNU General Public License (GPL).
If you are using PhyloSort in a published work or product, please cite: Ahmed Moustafa and Debashish Bhattacharya. PhyloSort: A user-friendly phylogenetic sorting tool and its application to estimating the cyanobacterial contribution to the nuclear genome of Chlamydomonas. BMC Evol. Biol. 8:6 (open access) (Qualified as highly accessed).
Acknowledgments
- National Institutes of Health (NIH) for supporting our research.
- ej-technologies for providing a license for install4j to generate the multi-platform installers.